Backend For Reasoning About Interaction Networks
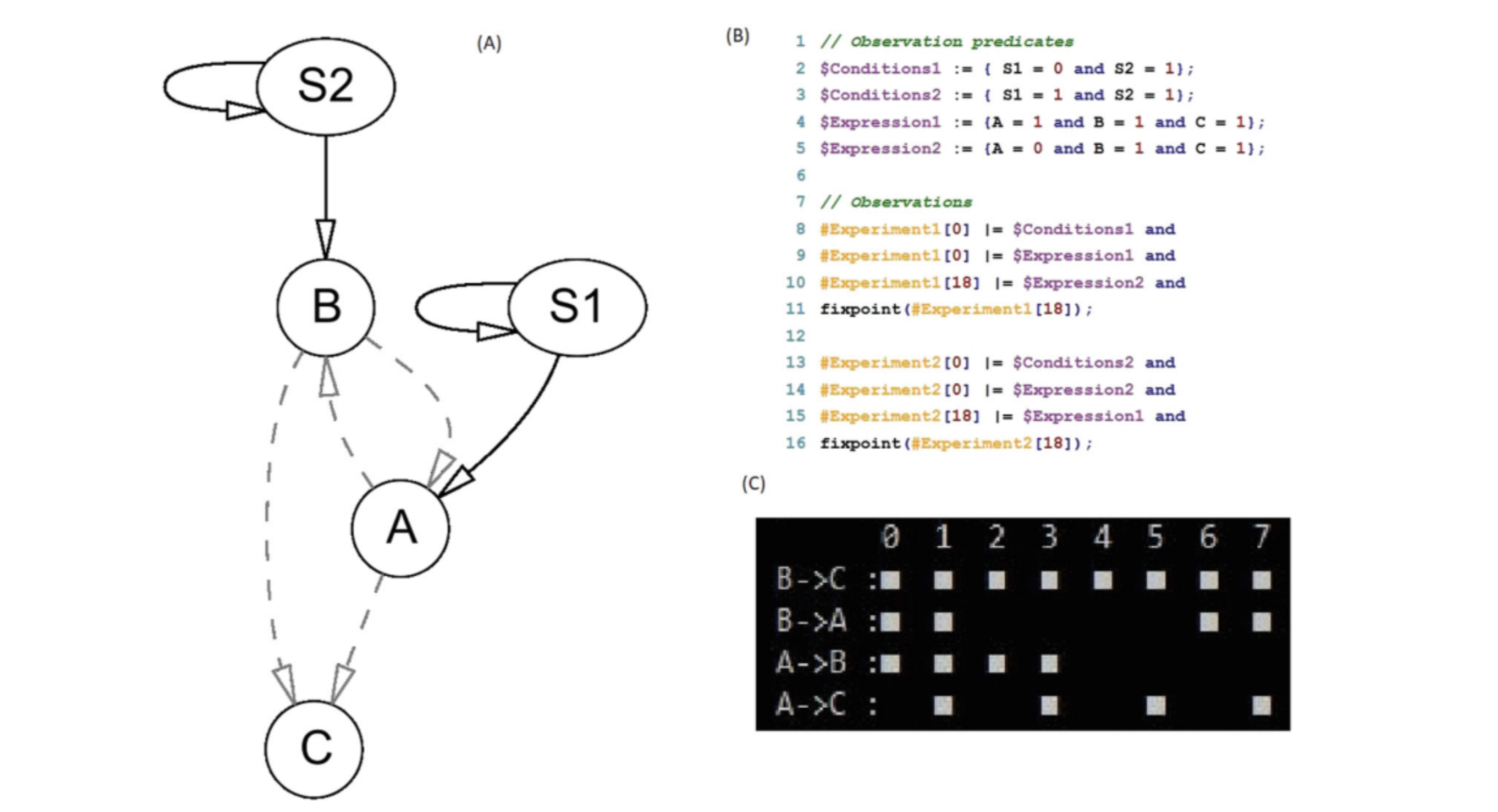
In the summer of 2018, I began working with Hillel Kugler on implementing an open source version of RE:IN, a tool developed by Micrososft Research to analyze observations about cell states, and conclude what gene interactions were and were not consistent woth the observations.
Eventually, my implementation was able to be competitive with, and in some cases surpass, RE:IN on some benchmarks, and my implementation also had some additional capabilities in terms of what sorts of relationships it was able to express. This work led to the publication of two papers:
For more details, checkout the github.
Although I am no longer actively involved, my work is still being extended and built upon, as can be seen here.
